Below the fold, some details and a little discussion.
Up to now the UW/Microsoft team have used Illumina's "sequencing by synthesis" (SBS) as the technology for reading the sequence of bases from the DNA strand. But this time they used:
Nanopore sequencing, as commercialized by Oxford Nanopore Technologies (ONT), offers a sequencing alternative that is portable, inexpensive and automation-friendly, resulting in a better option for a real-time read-head of a molecular storage system. Specifically, ONT MinION is a four-inch long USB-powered device containing an array of 512 sensors, each connected to four biological nanopores, ... Each nanopore is built into an electrically resistant artificial membrane. During sequencing, a single strand of DNA passes through the pore resulting in a change in the current across the membrane. This electrical signal is processed in real time to determine the sequence identity of the DNA strand.Nanopore has two key advantages over SBS for reading data from DNA:
In the context of DNA storage, real-time sequencing enables the ability to sequence until sufficient coverage has been acquired for successful decoding without having to wait for an entire sequencing run to be completed. Moreover, nanopore sequencing offers a clear single-device throughput scalability roadmap via increased pore count, which is very important for viability of DNA data storage.But there are significant problems too:
Nanopore sequencing presents unique challenges to decoding information stored in synthetic DNA. In addition to a significantly higher error rate compared to SBS, nanopore sequencing of short DNA fragments results in lower sequencing throughput due to inefficient pore utilization. ... existing scalable approaches for writing synthetic DNA rely on parallel synthesis of millions of short oligonucleotides (i.e., 100–200 bases in length) where each oligonucleotide contains a fraction of an encoded digital file. We find that sequencing of such short fragments results in significantly lower sequencing throughput in the ONT MinION. This limitation hinders the scalability of nanopore sequencing for DNA storage applications.So, for nanopore to be effective, the data needs to be encoded in much longer strands. The detailed reason is that:
The overall yield and quality of a nanopore sequencing run is dependent on the molecular size of the DNA to be sequenced. DNA molecules translocate through the pore at a rate of 450 bases/sec while it can take between 2–4 s for a pore to capture and be occupied by the next DNA molecule. Therefore, short DNA molecules result in a higher number of unoccupied pores over time, which increases the rate of electrolyte utilization above the membrane. This results in a faster loss in polarity and lower sequencing capacity and overall throughput. ONT estimates that the optimal DNA size to maximize sequencing yield is around 8 kilobases while the minimum size is 200 bases. Below 200 bases, event detection and basecalling is not possible.Their approach is to concatenate many of their short strands:
To achieve this, we implement a strategy that enables both random access and molecular assembly of a given DNA file stored in short oligonucleotides (150 bp) into large DNA fragments containing up to 24 oligonucleotides (~5000 bp). We evaluate Gibson Assembly and Overlap-Extension Polymerase Chain Reaction (OE-PCR) as suitable alternatives to iteratively concatenate and amplify multiple oligonucleotides in order to generate large sequencing reads.They preferred Gibson Assembly for this task. The remaining problem is the high error rate:
To decode the files, we implement a consensus algorithm capable of handling high error rates associated with nanopore sequencing.
 |
| Figure 3a |
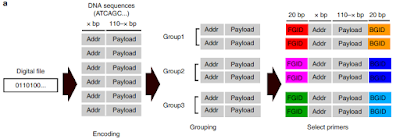
Each file in the oligo pool consists of a set of 150-bases oligonucleotides with unique 20-nucleotide sequences at their 5′ and 3′ ends for PCR-based random-access retrieval (i.e., file ID) and a 110-nucleotide payload encoding the digital information.The system repeatedly reads many copies of these strands (at least 36 in their previous work) and applies a decoding algorithm to the resulting base sequences to identify and correct errors:
In our previous decoding algorithm, the consensus sequence is recovered by a process where pointers for payload sequences are maintained and moved from left to right, and at every stage of the process the next symbol of the sequence is estimated via a plurality vote. For payload sequences that agree with plurality, the pointer is moved to the right by 1. But for the sequences that do not agree with plurality, the algorithm classifies whether the reason for the disagreement is a single deletion, an insertion, or a substitution. This is done by looking at the context around the symbol under consideration. Once this is estimated, the pointers are then moved to the right accordingly.Nanopore has a higher error rate than SBS, so this algorithm would need even more than 36 copies. By improving the algorithm they instead reduced the minimum number of copies to 22:
The key difference between our new algorithm and our previous implementation is that in cases when disagreements cannot be classified, we do not drop respective payload sequences from further consideration. Instead, we label such payload sequences as being out of sync and attempt to bring them back at later stages. Specifically, every sequence that is out of sync is ignored for several next steps of the algorithm. However, after those steps, we perform search for a match between the last few bases of the partially constructed consensus sequence and appropriately located short substrings of the payload sequence.One pore can decode a 5000bp strand in 11s, plus 2-4s recovery time. Lets say 14s. That's 308s for 22 reads, which will generate consensus on 2640 bases of data. At the theoretical maximum of 2bit/base this is 5280 bits of data. So the read bandwidth per pore is about 17bit/s.
If we discover a match, we move the payload sequence pointer to the corresponding location and drop the out of sync label. This allows the payload sequence pointer to circumvent small groups of adjacent incorrect bases, a feature that was not present in the earlier algorithm. This modification to the consensus algorithm allows us to successfully decode from notably lower coverages because more information from the sequencing reads is used in the process.
For example, Seagate's Exos 7E8 drives have a sustained transfer rate of 215Mb/s. To match the read performance of a single hard drive would need about 13M pores, or about $6.2M worth of MiniONs. (ONT does have higher-throughput products, using the PromethION48 would need 88 units at about $52M). This doesn't account for Reed-Solomon overhead, or the difficulty of ensuring that each strand was read only 22 times among the 13M pores. Although this statement is accurate:
nanopore sequencing offers a clear single-device throughput scalability roadmap via increased pore count, which is very important for viability of DNA data storage.it rather understates the engineering difficulties involved in scaling up to match competing media.
No comments:
Post a Comment